Diferenciální diagnostika glioblastomu a solitárních metastáz mozku – úspěch modelů umělé inteligence vytvořených na základě radiomických dat získaných automatickou segmentací z konvenčních MR sekvencí
Diferenciální diagnostika glioblastomu a solitárních metastáz mozku – úspěch modelů umělé inteligence vytvořených na základě radiomických dat získaných automatickou segmentací z konvenčních MR sekvencí
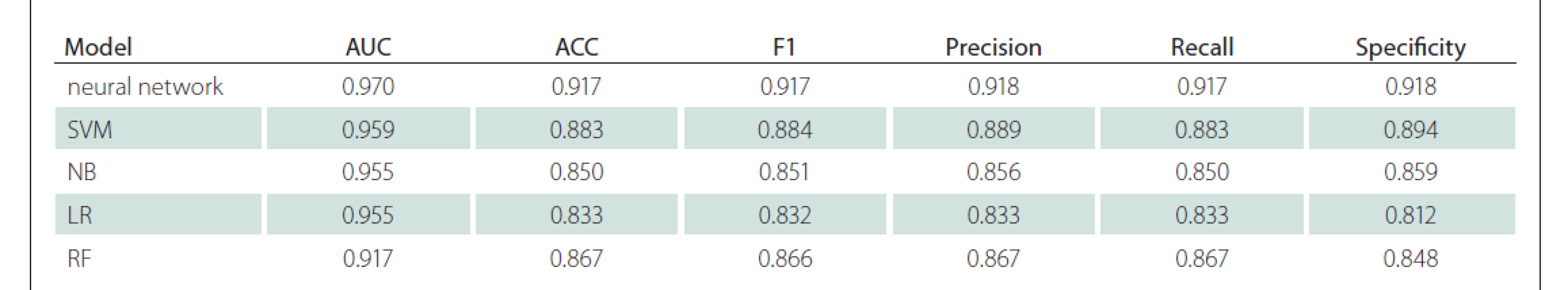
Cíl: Cílem naší studie bylo odlišit glioblastom (GBM) od solitární metastázy mozku za pomoci strojových modelů vyvinutých na základě radiomických dat získaných automatickou segmentací nádoru z konvenčích MR skenů pacientů pomocí umělé inteligence. Metody: Naše studie byla prováděna na jednom pracovišti a byla retrospektivní. Do studie bylo zařazeno 35 pacientů s GBM a 25 pacientů se solitární metastázou na mozku, u nichž byla před operací provedena MR mozku s kontrastní látkou. Do programu BraTumIA byly nahrány T1 vážené obrazy, T1 vážené obrazy po podání kontrastní látky, T2 vážené obrazy a T2 vážené obrazy s využitím sekvence fluid attenuated inversion recovery (FLAIR). V programu byly léze pacienta pomocí umělé inteligence rozděleny do čtyř různých segmentů: nekróza, nesytící se solidní oblast, sytící se solidní oblast a peritumorózní edém. Z T1 obrazů po podání kontrastní látky a T2 FLAIR obrazů bylo extrahováno 856 znaků. Pro výběr znaků, optimalizaci modelu a validaci byl použit vnořený (nested) přístup. Byly modelovány umělé neuronové sítě, podpůrný vektorový stroj, náhodný les a naivní bayesovský klasifikátor. Funkce modelu byla hodnocena pomocí přesnosti, senzitivity, specificity a plochy pod křivkou (area under the curve; AUC). Výsledky: Mezi skupinami s GBM a s metastázou nebyly rozdíly ve věku a pohlaví. Nejúspěšnější výsledky byly získány pomocí algoritmu neuronové sítě – byla získána hodnota AUC 0,970. U algoritmů za použití podpůrného vektorové stroje, naivního bayesovského klasifikátoru, logistické regrese či náhodného lesu byly získány hodnoty AUC 0,959, 0,955, 0,955, respektive 0,917. Závěr: V diferenciální diagnostice GBM a solitárních metastáz mozku mohou modely umělé inteligence založené na radiomických datech pomocí automatické segmentace objektivně a s vysokou přesností odlišovat tak, že závislost na prostředku a osobě udržují na nejnižší úrovni za použití prostých konvenčních sekvencí.
Klíčová slova:
radiomika – strojové učení – glioblastoma – metastazující nádor na mozku – analýza textury – automatická segmentace
Authors:
E. Demirel 1; C. O. Gökaslan 1; O. Dilek 2; C. Ozdemir 3; M. G. Boyacı 4; S. Korkmaz 4
Authors‘ workplace:
Department of Radiology, Afyonkarahisar Health Sciences University, Afyonkarahisar, Turkey
1; Department of Radiology, University of Health Sciences, Adana Teaching and Research Hospital, Adana, Turkey
2; Department of Pathology, Afyonkarahisar Health Sciences University, Afyonkarahisar, Turkey
3; Department of Neurosurgery, Afyonkarahisar Health Sciences University, Afyonkarahisar, Turkey
4
Published in:
Cesk Slov Neurol N 2021; 84(6): 541-546
Category:
Original Paper
doi:
https://doi.org/10.48095/cccsnn2021541
Overview
Aim: Our study aimed to distinguish glioblastoma (GBM) from solitary brain metastasis with machine models developed with radiomics data obtained by artificial intelligence-based automatic tumour segmentation over conventional MRI of the patients. Methods: Our study was conducted as single-centre and retrospective. Thirty-five GBM and 25 solitary brain metastasis patients who had pre-operative contrast-enhanced brain MRI were included in the study. T1-weighted, postcontrast T1-weighted, T2-weighted and fluid attenuated inversion recovery (FLAIR) T2-weighted images of the patients were uploaded to the program named BraTumIA. With the program, the patient‘s lesions were divided into four different segments by artificial intelligence as necrosis, non-enhancing solid area, enhancing solid area and peritumorous oedema. 856 features were extracted from T1 post-contrast and T2 FLAIR images. A nested approach was used for feature selection, model optimization and validation. Artificial neural networks, support vector machine, random forest and naive bayes were modelled. Accuracy, sensitivity, specificity and area under the curve (AUC) parameters were used to evaluate the model performance. Results: There was no difference between GBM and metastasis groups in terms of age and gender. The most successful results were obtained in the neural network algorithm; 0.970 AUC was found. Other support vector machine, naive bayes, logistic regression and random forest algorithms also found 0.959, 0.955, 0.955, 0.917 AUC values, respectively. Conclusion: In the differential diagnosis of GBM and solitary brain metastasis, radiomics-based artificial intelligence models obtained by automatic segmentation can distinguish objectively and with high accuracy by keeping device and person dependency at the lowest level with only conventional sequences.
Keywords:
machine learning – glioblastoma – radiomics – metastatic brain tumour – texture analysis – automatic segmentation
Introduction
Metastases and glioblastomas (GBM) are the most common malignant lesions in the brain in adulthood [1]. In conventional brain MRI, GBM and metastases have a similar imaging pattern [2]. Since these two lesions‘ treatment processes are completely different from each other, it is essential to differentiate these two lesions by non-invasive methods. Currently, the gold standard for diagnosing brain tumours is based on histopathology [3] However, brain tumour biopsy-related complications occur in about 6% of biopsy cases [4].
MRI techniques have demonstrated advanced innovations in recent years. Studies with innovative techniques such as MR perfusion, MR spectroscopy (MRS) and diffusion tensor ımaging (DTI) are increasing day by day in order to distinguish these two lesions from each other and very successful results have been obtained [5–8]. However, prolonged acquisition times, long post-processing processes, the lack of active modalities in every available device and the ability to be added to devices only with very high fees make the applicability of these modalities very difficult to be used in every case.
Radiomics can be defined as a system that extracts high-throughout quantitative features from radiographic images and provides quantitative data far beyond what the eye can see [9]. With radiomics, it is possible to extract more quantitative features than can be distinguished from conventional routine sequences.
In our study, we aimed to distinguish between GBM and solitary brain metastasis with machine models developed on radiomics data obtained by artificial intelligence-based automatic tumour segmentation using conventional MRI of the patients.
Material and methods
Patient selection
Thirty-five GBM and 25 solitary brain metastasis patients (from January 2011 to June 2020) from Afyonkarahisar Health Sciences University Centre were included. Inclusion criteria were as follows: patients with pre-operative imaging; patients with complete conventional sequences (T1-weighted images [T1WI], T2-weighted images [T2WI], T2 fluid attenuated inversion recovery [FLAIR], postcontrast T1WI), patients with clearly defined diagnoses of GBM and brain metastasis in the pathology report. The exclusion criteria were as follows: patients with multiple lesions, MRI images with obvious artifacts or inadequate imaging quality; patients with a history of intracranial disease (such as brain trauma, intracranial infection, other brain tumours).
One hundred and twenty-five patients who met the inclusion criteria were included in the study. Thirty of them were excluded because of multiple lesions, 6 patients due to inappropriate pathology reports and 29 of them due to artifacts and inadequate imaging quality (Fig. 1).
N – number
Obr. 1. Vývojový diagram výběru
pacienta.
N – počet

Magnetic resonance imaging
All patients underwent unenhanced brain MRI examination on a 1.5 Tesla Philips Intera (Philips Medical Systems; Amsterdam, Netherlands) and a 1.5 Tesla Siemens Aera (Siemens, Erlangen, Germany) scanner. T1 axial (field-of view [FOV]: 200 mm; matrix: 574 × 574; thickness: 2 mm; echo time [TE]: 4; 5 ms; time to repeat [TR]: 25 ms); T2 FLAIR (FOV: 200 mm; matrix: 574 × 574; thickness: 2 mm; TE: 105 ms; TR: 9,000 ms; inversion time: 2,500 ms), T2 (FOV: 200 mm; matrix: 574 × 574; thickness: 2 mm; TE: 140 ms; TR: 4,200) T1 post contrast images were evaluated. An overview of the radiomics workflow is shown in Fig. 2.
GLCM – gray level co-occurrence matrix; GLRLM – gray level run length matrix; GLSZM – gray level size zone matrix; NGTDM – neighbourhood
gray-tone difference matrix
Obr. 2. Pracovní postup technické studie.
GLCM – úroveň šedé matice současného výskytu; GLRLM – úroveň šedé matice délky trvání; GLSZM – úroveň šedé matice velikosti zóny;
NGTDM – matice rozdílu šedé v sousedství

Segmentation
For fully automated segmentation, the open-source program Brain Tumor Image Analysis (BraTumIA) v.2.0.5 (NeuroImaging Tools and Resources Collaboratory) was used (Fig. 3) [10]. The program requires the user to upload conventional MRI sequences (T1WI, post-contrast T1WI, T2WI and FLAIR) into the program interface. The system uses four different MRI sequences to identify the tumour and its subregions. BraTumIA not only separates the lesion from the intact brain tissue but also performs segmentation of tumour tissue into four sub-regions (oedema, necrosis, non-enhancing solid area and enhancing solid area). Initially, images pass through a pre-processing line that includes aligning the images, extracting brain tissue from the images and extracting noise from the signal. Then, the feature extraction process, which includes the identification of different properties for each voxel, is applied to pre-processed images to distinguish between pathological and healthy tissue. Classification is done using a Support Vector Machine classifier, which determines, based on each voxel‘s characteristics, the allocation of it to one of the subdivisions evaluated through a probability distribution. Finally, using a Conditional Random Fields method on the created tag map, the spatial arrangement is tried to ensure the spatial consistency of voxels classified according to neighbouring voxels. This whole pipeline of the program has been described before [11,12]. Subsequently, minor segmentation flaws were corrected manually with the 3D slicer v4.11 program.
Extraction of radiomics features
The volume-of-interest was then normalized by the package “NormalizeImageFilter”. Before feature extraction by the 3DSlicer version 4.11 radiomic package (version 2.1.0), gray-level discretization and voxel resampling were performed. All features were calculated with a fixed bin width of 25, and resampling to a voxel size of 0.6 x 0.6 x
FLAIR – fluid-attenuated inversion recovery;
T1 C+ – enhanced T1; WI – weighted images
Obr. 3. Uživatelské rozhraní programu
BraTumIA. Program BraTumIA masu rozděluje
do 4 segmentů: červená – nekróza;
modrá – edém; růžová – solidní oblast bez
sycení po kontrastu; žlutá – solidní oblast
sytící se po kontrastu.
FLAIR – inverzní zobrazení s potlačením signálu
tekutiny; T1 C+ - T1 s kontrastem;
WI – vážené obrazy

Dimension reduction and machine learning
The open-source Python Sci-Kit Learn library and Waikato Environment for Knowledge Analysis toolkit version 3.8.4 (The University of Waikato, Hamilton, New Zealand) were used [16,17]. It is crucial to remove data that do not contribute significantly to the classification‘s accuracy from the entire feature set and create a model with contributing data. For this reason, the attributes went through certain preliminary processes before machine learning was introduced. First, randomization and normalization of the features were provided. Collinearity analysis was performed using the Pearson‘s correlation coefficient (r) test. The r threshold was 0.7. The features with high collinearity were excluded from the analysis. We used a nested cross-validation method (5-fold inner and 5-fold outer cross-validation loops) for feature selection along with model optimization. For feature selection, a wrapper attribute evaluator and an incremental wrapper-based subset selection method were used [18–20]. The wrapper attribute evaluator evaluates attribute sets of interest by user-defined learning schemes, which were support vector machine (SVM) in our study. The attributes that underwent two or more inner loops were included in the outer loop. Then, the features that underwent two or more outer cross-validations were chosen for final model development.
In machine learning, artificial neural networks (ANN), SVM, logistic regression (LR) random forest (RF) and naive bayes (NB) were modelled. Due to the relatively small size of our data set, the nested cross-validation method was preferred. Nested cross-validation was performed via a 5-fold outer and inner loop. The performance of classifiers was mainly evaluated and compared with the area under the curve (AUC).
Statistical analysis
Descriptive statistics; mean ± standard deviation for continuous variables if it conforms to the normal distribution; if it did not comply with the normal distribution, it was expressed as median values. The Mann-Whitney U test was used to compare continuous variables that did not show normal distribution with two-level variables. Relationships between categorical variables were examined by Chi-square analysis / Fisher‘s exact test. Finally, the relationships between continuous variables were evaluated using Pearson correlation analysis. The significance level was accepted as P < 0.05 in all analyzes. The open-source R program was used for these processes.
Results
A total of 60 patients were included in the study, 38 males and 22 females, aged between 28 and 94 years. Thirty-five patients were diagnosed with GBM and 24 had metastasis. Primary diagnoses of metastatic brain tumours were as follows; lung carcinoma (N = 12), colorectal carcinoma (N = 5), breast cancer (N = 3), renal cell carcinoma (N = 2), malignant melanoma (N = 1) and cancer of unknown primary (N = 1). There were no significant differences in sex and age between the two groups (P = 0.651, P = 0.910, respectively).
As a result of the dimension reduction processes, 280 features were eliminated due to the collinearity analysis. Then, as a result of wrapper subset-based nested cross-validation processes, machine models were fed with four attributes containing the most information.
When the successes in machine learning were ranked according to the AUC value, the highest success was found in ANN. It was able to differentiate GBM/metastasis with a 0.970 AUC value, 92% sensitivity and 92% specificity. Subsequently, the SVM, NB, LR, RF models had AUC success, respectively. Detailed classification performance parameters are available in Tab. 1, and the models‘ receiver operating characteristic curves are shown in Fig. 4.
SVM – support vector machine
Obr. 5. ROC křivka optimálního klasifikátoru.
SVM – podpůrný vektorový stroj

Discussion
In our study, we aimed to distinguish GBM from solitary brain metastasis with machine models developed with radiomics data obtained by artificial intelligence-based automatic tumour segmentation through only conventional MRI images of the patients. In our working system, we aimed to work as objectively as possible by minimizing individual dependency, including the segmentation process. We have achieved very high success in distinguishing GBM from solitary brain metastasis. However, since our study group is relatively small, our study can be considered as a pilot study and more robust models should be developed with larger patient series.
Differential diagnosis of GBM and metastasis, which are the most common malignant tumours of the brain, is quite challenging, especially in the presence of a solitary lesion, if there is no proven malignancy. Considering studies in the literature, advanced MRI methods such as MRS, MR perfusion and DTG can successfully differentiate GBM from solitary metastases [5–8]. However, it is challenging to use all advanced MRI modalities in the first examination for tumour imaging due to human and device-based limitations.
Radiomics refers to a process that extracts high-throughout quantitative and objective features from radiographic images and creates image features prediction models for genomic patterns and clinical outcomes [9]. While obtaining these features, segmentation of the lesion or organ constitutes a vital problem that creates user dependency. Many radiomics-based studies in current literature partially lose their objectivity due to segmentation with manual segmentation [21]. Although researchers segment with more than one person and exclude discordant data between evaluators, this situation is quite time-consuming. Therefore, the number of studies involving automatic segmentation is increasing day by day in studies based on radiomics [22]. This situation is quite challenging for researchers due to the need for advanced engineering knowledge and the lack of user-friendly open-source third-party software. In our study, we managed to partially overcome this problem with the completely free and user-friendly BraTumIA program.
Since our inclusion/exclusion criteria were quite limiting, we had difficulty in expanding our sample group. In particular, pathologically proven solitary metastasis was our main limitation. Although there are public datasets for GBM, there are no such data for solitary brain metastases. We preferred a nested cross-validation approach to partially overcome the overfitting and selection bias problems that our working group‘s small size may cause.
There are few radiomics-based studies for the differential diagnosis of GBM and metastasis. Bae et al [23] extracted radiomics data on the enhancing area and peritumorous oedema area that they segmented semiautomatically in sample groups that included 166 training and 82 verification cohorts. They succeeded in making this distinction in the deep neural network with a value of 0.956 AUC in the artificial intelligence models they developed. Chen et al [24] succeeded in making this discrimination with an AUC value of 0.830 in their artificial intelligence models‘ logistic regression model. They extracted radiomics data from the enhancing tumour area in the manual segment in 134 disease sample groups. Ortiz-Ramón et al [25] obtained radiomics data by manually segmenting postcontrast T1WI from a sample group of 50 GBM and 50 metastases. As in our study, they used the nested cross-validation method. Again, in the artificial intelligence models they developed, they found a value of 0.896 AUC in support vector machines. Qian et al [26] found a value of 0.900 AUC in support vector machines in artificial intelligence models that they developed by extracting radiomics data from the manually segmented enhancing tumour area in the sample group consisting of 242 GBM and 170 metastases.
Artzi et al [27] performed the most comprehensive study on this subject with a sample group of 212 GBM and 227 metastasis patients. The patients‘ lesions were semi-automatically segmented on postcontrast T1WI, which is the superiority of this study to previous studies in the literature. However, there is partial user dependency in the semiautomatic segmentation process they used. In the artificial intelligence models, they found a value of 0.940 AUC in support vector machines.
When evaluated together with previous studies, we achieved very high success in almost all of the machine models we developed. In the ANN algorithm, we distinguished GBM/metastasis with 0.970 AUC value, 92% sensitivity and 92% specificity. The most significant advantage of our work is that it is carried out using open-source programs. It is the only study in the literature to segment the mass as necrosis, non-enhancing solid area, enhancing solid area and peritumourous oedema area with fully automated segmentation and extracting radiomics data separately in these four areas. In addition, it is another advantage that it obtains radiomics data over both post-contrast T1WI and FLAIR.
The tests used in this differential diagnosis must have similar predictive values and confidence intervals in diseases with relatively common and catastrophic consequences in the population, such as GBM and metastasis. Models created with radiomics data are still far behind this generalisability. When it comes to MRI, many parameters such as Tesla differences, vendor differences and differences in the obtained sequence parameters change the image‘s quantitative properties before the radiomics features extraction process. Later, it contributes to this difference in many stages such as pre-processing applications, radiomics feature extraction applications, feature selection methods, machine learning algorithms and hyperparameter tuning processes. Our study has followed an approach that will try to increase generalisability using a 1.5 T machine from two different vendors and with as few features as possible. For the models created with radiomics data to gain generalisability, a robust feeding model should be selected and systems should be fed with the images of the maximum number of patients from the most different devices possible. For this reason, our study is only a pilot study.
When the supplementary packages of the studies were examined, it was seen that the most frequently used features in model development were “GLSZM nonuniformity” and size-related parameters [24,28]. In our study, shape-related features and GLSZM nonuniformity were among the parameters used in model development. If this situation is considered in future studies, fast and accurate systems can be created with much fewer features.
There are some very important limitations in our study. The most important of these is that it is a single-centre retrospective and a small sample group. Due to a small sample group, patients could not be allocated for the external validation cohort. Although the nested cross-validation approach was chosen to overcome this, new studies based on automated segmentation with larger validation cohorts will yield more reliable and generalisable results. Since there are many different primary malignancies that cause heterogeneity of the metastasis group, more information can be obtained if the number of metastasis cases is increased and divided into subgroups.
Conclusion
As a result, our study is unique in the literature due to the application of automatic segmentation process with completely free useful programs and the extraction of radiomics data from 4 different regions of the mass through postcontrast T1WI and T2 FLAIR images, which are the most important sequences in conventional tumour imaging. With the widespread use of automatic segmentation and radiomics applications over time, robust-fed machine models with minimum user dependency and high generalisability will serve as a successful assistant in the decision-making process in radiology.
Ethical principles
The entire study was conducted in accordance with the Helsinki Declaration of 1975 (as revised in 2004 and 2008). The Institutional Review Board approved this retrospective study and all subjects provided written informed consent (Ethical Committee-2020/367).
Conflict of interest
The authors declare they have no potential conflicts of interest concerning drugs, products or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE “uniform requirements” for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do biomedicínských časopisů.
Okan Dilek, MD
Department of Radiology
University of Health Sciences
Adana Teaching and Research
Hospital
Dr. Mithat Özsan Bulvarı Kışla Mah.
4522 Sok. No: 1 Yüreğir
Adana, Turkey
e-mail: dr.okandilek@gmail.com
Accepted for review: 24. 5. 2021
Accepted for print: 8. 11. 2021
Sources
1. Lemke DM. Epidemiology, diagnosis, and treatment of patients with metastatic cancer and high-grade gliomas of the central nervous system. J Infus Nurs 2004; 27 (4): 263–269. doi: 10.1097/00129804-200407000-00012.
2. Maurer MH, Synowitz M, Badakshi H et al. Glioblastoma multiforme versus solitary supratentorial brain metastasis: differentiation based on morphology and magnetic resonance signal characteristics. Rofo 2013; 185 (3): 235–240. doi: 10.1055/s-0032-1330318.
3. Pollo B. Pathological classification of brain tumors. Q J Nucl Med Mol Imaging 2012; 56 (2): 103–111.
4. Malone H, Yang J, Hershman DL et al. Complications following stereotactic needle biopsy of intracranial tumors. World Neurosurg 2015; 84 (4): 1084–1089. doi: 10.1016/j.wneu.2015.05.025.
5. Blasel S, Jurcoane A, Franz K et al. Elevated peritumoural rCBV values as a mean to differentiate metastases from high-grade gliomas. Acta Neurochir 2010; 152 (11): 1893–1899. doi: 10.1007/s00701-010-0774-7.
6. Server A, Josefsen R, Kulle B et al. Proton magnetic resonance spectroscopy in the distinction of high-grade cerebral gliomas from single metastatic brain tumors. Acta Radiol 2010; 51 (3): 316–325. doi: 10.3109/02841850903482901.
7. Wang S, Kim S, Poptani H et al. Diagnostic utility of diffusion tensor imaging in differentiating glioblastomas from brain metastases. AJNR Am J Neuroradiol 2014; 35 (5): 928–934. doi: 10.3174/ajnr.A3871.
8. Jung BC, Arevalo-Perez J, Lyo JK et al. Comparison of glioblastomas and brain metastases using dynamic contrast-enhanced perfusion MRI. J Neuroimaging 2016; 26 (2): 240–246. doi: 10.1111/jon.12281.
9. Lambin P, Rios-Velazquez E, Leijenaar R et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48 (4): 441–446. doi: 10.1016/j.ejca.2011.11.036.
10. Porz N, Bauer S, Pica A et al. Multi-modal glioblastoma segmentation: man versus machine. PloS One 2014; 9 (5): e96873. doi: 10.1371/journal.pone.0096873.
11. Rios Velazquez E, Meier R, Dunn WD et al. Fully automatic GBM segmentation in the TCGA-GBM dataset: prognosis and correlation with VASARI features. Sci Rep 2015; 5: 16822. doi: 10.1038/srep16822.
12. Porz N, Habegger S, Meier R et al. Fully Automated enhanced tumor compartmentalization: man vs. machine reloaded. PLoS One 2016; 11 (11): e0165302. doi: 10.1371/journal.pone.0165302.
13. Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. IEEE Trans Syst Man Cybern 1973; 3 (6): 610–621.
14. Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process 1975; 4 (2): 172–179. doi: 10.1016/S0146-664X (75) 80008-6.
15. Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit Lett 1990; 11 (6): 415–419. doi: 10.1016/0167-8655 (90) 90112-F.
16. Abraham A, Pedregosa F, Eickenberg M et al. Machine learning for neuroimaging with scikit-learn. Front Neuroinform 2014; 8: 14. doi: 10.3389/fninf.2014.00014.
17. Hall M, Frank E, Holmes G et al. The WEKA data mining software: an update. SIGKDD Explor 2009; 11 (1): 10–18. doi: 10.1145/1656274.1656278.
18. Kohavi R, John GH. Wrappers for feature subset selection. Artif Intell 1997; 97 (1–2): 273–324. doi: 10.1016/S0004-3702 (97) 00043-X.
19. Su X, Sun H, Chen N et al. A radiomics-clinical nomogram for preoperative prediction of IDH1 mutation in primary glioblastoma multiforme. Clin Radiol 2020; 75 (12): 963.e7–963.e15. doi: 10.1016/j.crad.2020.07. 036.
20. Suh HB, Choi YS, Bae S et al. Primary central nervous system lymphoma and atypical glioblastoma: differentiation using radiomics approach. Eur Radiol 2018; 28 (9): 3832–3839. doi: 10.1007/s00330-018-5368-4.
21. Haarburger C, Müller-Franzes G, Weninger L et al. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci Rep 2020; 10 (1): 12688. doi: 10.1038/s41598-020-69534-6.
22. Chen W, Liu B, Peng S et al. Computer-aided grading of gliomas combining automatic segmentation and radiomics. Int J Biomed Imaging 2018; 2018: 2512037. doi: 10.1155/2018/2512037.
23. Bae S, An C, Ahn SS et al. Robust performance of deep learning for distinguishing glioblastoma from single brain metastasis using radiomic features: model development and validation. Sci Rep 2020; 10 (1): 12110. doi: 10.1038/s41598-020-68980-6.
24. Chen C, Ou X, Wang J et al. Radiomics-based machine learning in differentiation between glioblastoma and metastatic brain tumors. Front Oncol 2019; 9: 806. doi: 10.3389/fonc.2019.00806.
25. Ortiz-Ramón R, Ruiz-España S, Mollá-Olmos E et al. Glioblastomas and brain metastases differentiation fol- lowing an MRI texture analysis-based radiomics approach. Phys Med 2020; 76: 44–54. doi: 10.1016/j.ejmp.2020.06.016.
26. Qian Z, Li Y, Wang Y et al. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 2019; 451: 128–135. doi: 10.1016/j.canlet.2019.02.054.
27. Artzi M, Bressler I, Ben Bashat D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging 2019; 50 (2): 519–528. doi: 10.1002/jmri.26643.
28. Dong F, Li Q, Jiang B et al. Differentiation of supratentorial single brain metastasis and glioblastoma by using peri-enhancing oedema region-derived radiomic features and multiple classifiers. Eur Radiol 2020; 30 (5): 3015–3022. doi: 10.1007/s00330-019-06460-w.
Labels
Paediatric neurology Neurosurgery NeurologyArticle was published in
Czech and Slovak Neurology and Neurosurgery

2021 Issue 6
Most read in this issue
- Stiff -person syndrom
- Normotenzní hydrocefalus
- Synukleinopatie a jejich laboratorní biomarkery
- Perorální kladribin v léčbě roztroušené sklerózy – data z celostátního registru ReMuS®
